-
Study
-
Quick Links
- Open Days & Events
- Real-World Learning
- Unlock Your Potential
- Tuition Fees, Funding & Scholarships
- Real World Learning
-
Undergraduate
- Application Guides
- UCAS Exhibitions
- Extended Degrees
- School & College Outreach
- Information for Parents
-
Postgraduate
- Application Guide
- Postgraduate Research Degrees
- Flexible Learning
- Change Direction
- Register your Interest
-
Student Life
- Students' Union
- The Hub - Student Blog
- Accommodation
- Northumbria Sport
- Support for Students
-
Learning Experience
- Real-World Learning
- Research-enriched learning
- Graduate Futures
- The Business Clinic
- Study Abroad
-
-
International
International
Northumbria’s global footprint touches every continent across the world, through our global partnerships across 17 institutions in 10 countries, to our 277,000 strong alumni community and 150 recruitment partners – we prepare our students for the challenges of tomorrow. Discover more about how to join Northumbria’s global family or our partnerships.
View our Global Footprint-
Quick Links
- Course Search
- Undergraduate Study
- Postgraduate Study
- Information for Parents
- London Campus
- Northumbria Pathway
- Cost of Living
- Sign up for Information
-
International Students
- Information for International Students
- Northumbria and your Country
- International Events
- Application Guide
- Entry Requirements and Education Country Agents
- Global Offices and Regional Teams
- English Requirements
- English Language Centre
- International student support
- Cost of Living
-
International Fees and Funding
- International Undergraduate Fees
- International Undergraduate Funding
- International Masters Fees
- International Masters Funding
- International Postgraduate Research Fees
- International Postgraduate Research Funding
- Useful Financial Information
-
International Partners
- Agent and Representatives Network
- Global Partnerships
- Global Community
-
International Mobility
- Study Abroad
-
-
Business
Business
The world is changing faster than ever before. The future is there to be won by organisations who find ways to turn today's possibilities into tomorrows competitive edge. In a connected world, collaboration can be the key to success.
More on our Business Services-
Business Quick Links
- Contact Us
- Business Events
- Research and Consultancy
- Education and Training
- Workforce Development Courses
- Join our mailing list
-
Education and Training
- Higher and Degree Apprenticeships
- Continuing Professional Development
- Apprenticeship Fees & Funding
- Apprenticeship FAQs
- How to Develop an Apprentice
- Apprenticeship Vacancies
- Enquire Now
-
Research and Consultancy
- Space
- Energy
- AI Futures
- CHASE: Centre for Health and Social Equity
- NESST
-
-
Research
Research
Northumbria is a research-rich, business-focused, professional university with a global reputation for academic quality. We conduct ground-breaking research that is responsive to the science & technology, health & well being, economic and social and arts & cultural needs for the communities
Discover more about our Research-
Quick Links
- Research Peaks of Excellence
- Academic Departments
- Research Staff
- Postgraduate Research Studentships
- Research Events
-
Research at Northumbria
- Interdisciplinary Research Themes
- Research Impact
- REF
- Partners and Collaborators
-
Support for Researchers
- Research and Innovation Services Staff
- Researcher Development and Training
- Ethics, Integrity, and Trusted Research
- University Library
- Vice Chancellors Fellows
-
Research Degrees
- Postgraduate Research Overview
- Doctoral Training Partnerships and Centres
- Academic Departments
-
Research Culture
- Research Culture
- Research Culture Action Plan
- Concordats and Commitments
-
-
About Us
-
About Northumbria
- Our Strategy
- Our Staff
- Our Schools
- Place and Partnerships
- Leadership & Governance
- University Services
- Northumbria History
- Contact us
- Online Shop
-
-
Alumni
Alumni
Northumbria University is renowned for the calibre of its business-ready graduates. Our alumni network has over 253,000 graduates based in 178 countries worldwide in a range of sectors, our alumni are making a real impact on the world.
Our Alumni - Work For Us
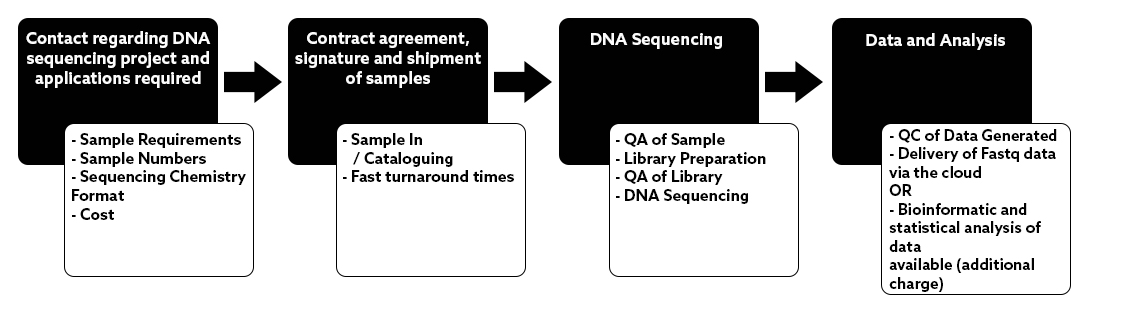
 NU-OMICS was amongst the first in the UK to install
the PacBio Sequel in 2017.
NU-OMICS was amongst the first in the UK to install
the PacBio Sequel in 2017.
This Single Molecule Real Time sequencing technology offers a powerful approach to de novo genome assemblies using long DNA sequencing reads with excellent consensus accuracy and uniform coverage.
Whole genome sequencing
- From simple to complex eukaryotes
- Providing excellent scaffolds for larger eukaryotes offering a cost effective approach to de novo sequencing
Small genome, in multiplex to reduce genome sequencing costs
- Currently multiplexing up to 15 bacterial genomes in one run. Higher density SMRT cells and longer movie chemistries will be available. Please ask for further details.
Complete 16S rRNA gene sequencing for Bacterial community analysis offering to species level comparison
- Using the following method to sequence the V1-V9 region of 16S rRNA gene
- Schloss PD, Jenior ML, Koumpouras CC, Westcott SL, Highlander SK. (2016) Sequencing 16S rRNA gene fragments using the PacBio SMRT DNA sequencing system. PeerJ 4:e1869 https://doi.org/10.7717/peerj.1869
Virus Genome sequencing
- Multiplexing available.
Methylation studies (prokaryotic/eukaryotic alongside reference)
Long transcript sequencing
- Eukaroyotes- Isoform sequencing
- Bacterial long transcript sequencing
We are always open to new research and projects. Contact us to discuss new approaches or implementing new workflows.
- As current kit based DNA extraction offers bias and shearing, gDNA preparations are best using phenol:chloroform extraction.
- All samples must be treated with broad spectrum RNAse prior to sending. An electrophoresis image is required for QA purposes run on a 1% agarose gel. Limited shearing should be present, if worried contact us to check before sending.
- All samples should be in a 96 well format as parts of the preparation are automated. Data will be returned labelled in well-format over sample name. DNA quality A260/280 should be >1.8.
- Sample numbers to be multiplexed determines the starting amount of DNA required. Please ask when being quoted for the project.
Please send samples to the following delivery address;
NU-OMICS DNA sequencing research facility
(C/O) Darren Smith/Andrew Nelson
Northumbria University
Ellison Building EBA 504a
Newcastle Upon Tyne
Tyne and Wear
NE1 8ST